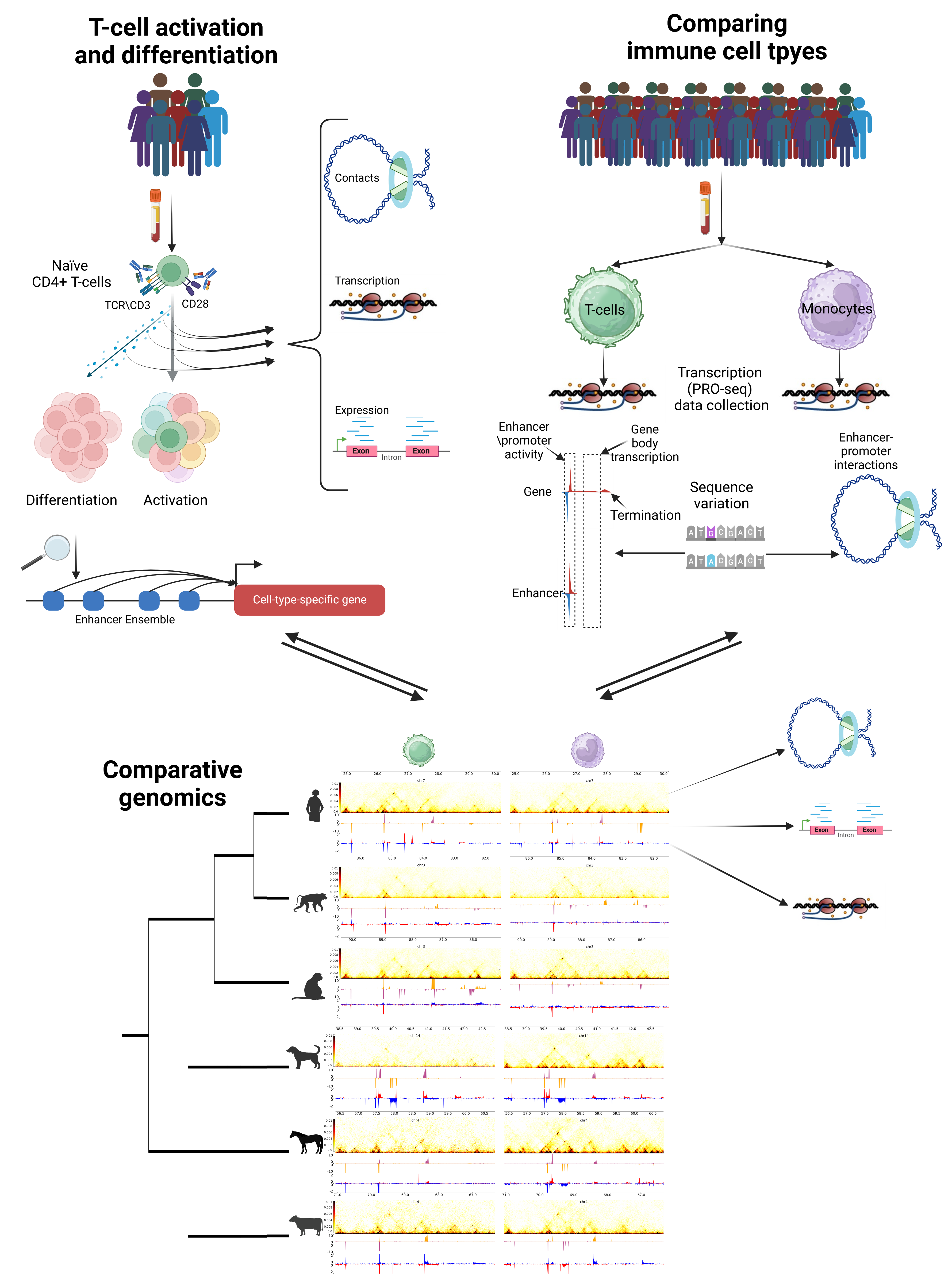
Our Focus
Complex biological traits, such as height, blood pressure, or susceptibility to various diseases, are defined by genetic interactions among multiple genomic elements and their interaction with environmental factors. Most sequence variants associated with complex traits are located within transcriptional enhancers—non-coding, often gene-distal regions of the genome. Enhancers are essential regulators, controlling when and where genes are activated, playing a crucial role in development and contributing to the biological complexity of multicellular organisms. Despite their immense importance, we have limited understanding of how enhancers transmit regulatory information over large genomic distances to their target genes. Therefore, deciphering how genetic variation in enhancers affects complex traits, including many human disorders, remains a central challenge in genomics and medicine.
In our lab, we aim to uncover the principles that define the effect of enhancer variation on complex traits. Currently, we focus on two major questions:
- What molecular and sequence determinants define the flow of regulatory information between enhancers and their target genes?
- Does the regulatory landscape of genes define the phenotypic effect of enhancer variants?
We use an array of genomic methods to measure nuclear properties such as genomic contacts, transcriptional activity and epigenetic marks, in developmental systems and in primary human cells, to understand how enhancers operate and how do they make us different.
Our Projects
Enhancer-Promoter Interactions and Transcriptional Regulation: What’s the Connection?
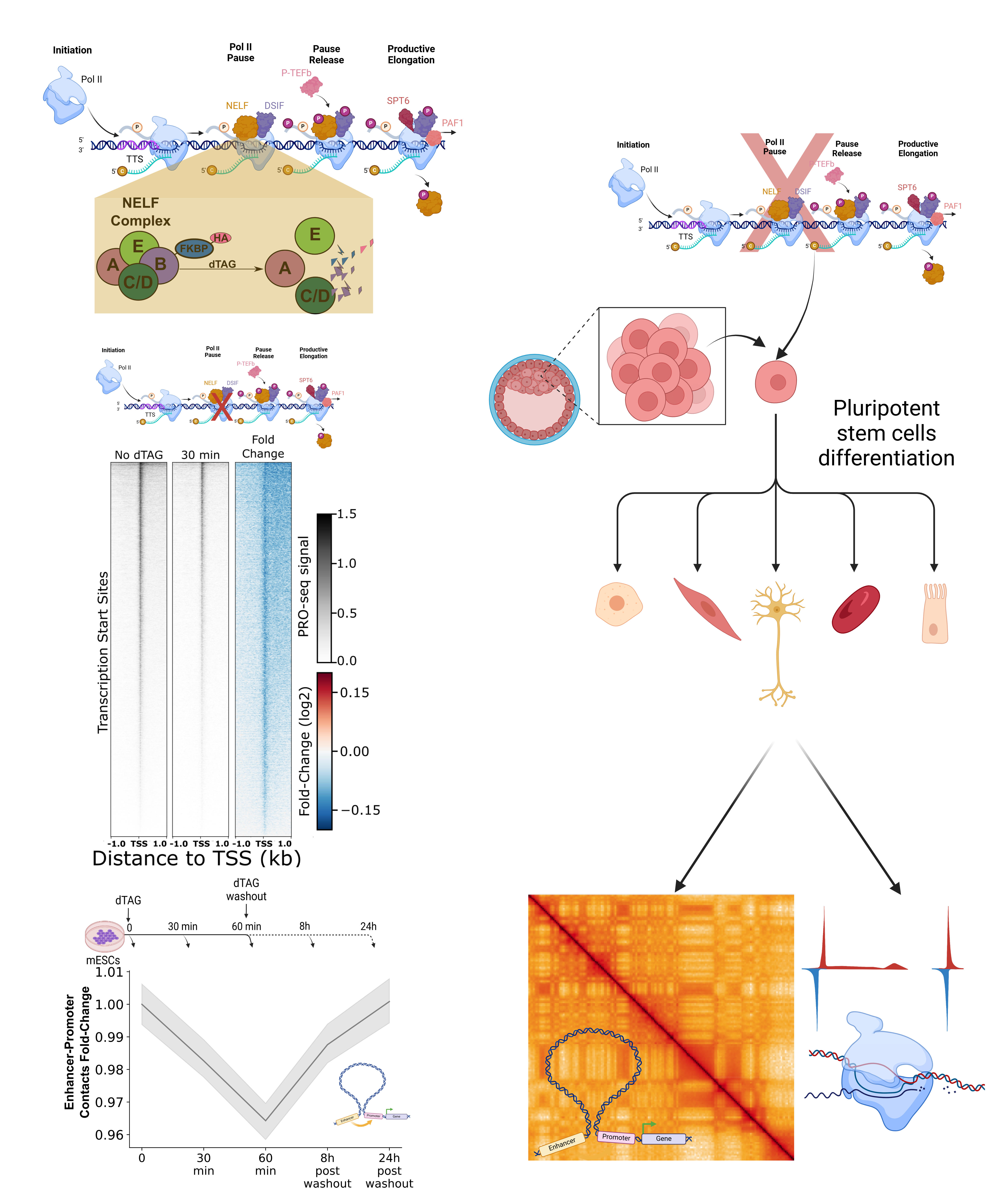
Enhancers control gene expression by regulating the rate of RNA synthesis (RNA transcription). However, enhancers are often located several kilobases to megabases away from the promoters of their target genes, creating a challenge for long-range communication of their regulatory information. Such long-range regulation requires two key components: a "glue" that brings enhancers and promoters together in space and "messenger" factors that transmit regulatory information from enhancers to their target promoters.
The transcription cycle in animals involves several crucial steps occurring near the transcription start site of a gene (or enhancer). These steps include chromatin opening, RNA polymerase recruitment, and transcription initiation. However, in most metazoan promoters and enhancers, RNA polymerase does not immediately transition from initiation to productive RNA synthesis. Instead, it pauses just downstream of the initiation site in a tightly regulated manner. Only after a series of phosphorylation events does the RNA polymerase proceed into productive elongation.
Until recently, the significance of this pausing step was not well understood. Our recent work demonstrated that RNA polymerase pausing in metazoans correlates with more frequent interactions between enhancers and promoters. This finding suggests a critical insight: enhancer-promoter interactions not only regulate transcription by RNA polymerase, but the dynamics of RNA polymerase itself also influence enhancer-promoter communication.
Leavraging dynamic in vitro differentiation systems, combined with directed targeting of factors in the transcriptional machinary, we strive to uncover how changes in key steps of the metazoan transcription cycle are shaped by—and, in turn, shape—enhancer-promoter communication.
Enhancer Variation and Phenotype: A Question of Context?
Enhancers are common targets of genetic variation. Many enhancer variants influence gene expression and are categorized as expression quantitative trait loci (eQTLs). However, a puzzling discrepancy exists: many enhancer eQTLs show no clear correlation with observable traits, while phenotypically relevant enhancer variants—such as those associated with diseases in genome-wide association studies (GWASs)—often fail to exhibit direct effects on gene expression. This disconnect complicates our ability to establish clear links between enhancer variation and complex phenotypes.
One key factor contributing to this discrepancy is the cellular context. Recent studies utilizing dynamic systems spanning multiple cell types, states, and conditions have revealed that some enhancer variants with phenotypic effects at the organismal level exhibit context-dependent effects on gene expression. These findings highlight the importance of considering cellular state and context when evaluating the regulatory impacts of enhancer variants.
Another critical factor is the regulatory landscape of target genes. Many enhancers operate within larger ensembles that regulate the same gene in a given cell type. This enhancer multiplicity can confer "phenotypic robustness" to changes in enhancer activity, meaning that alterations in individual enhancers may only manifest phenotypic consequences under specific stress conditions. Thus, to fully understand the effects of enhancer variation on gene expression and phenotype, it is essential to consider both the genomic context in which enhancers function and the cellular context in which their effects are observed.
Our research aims to unravel how genetic variation in enhancer sequences influences transcriptional regulation and enhancer-promoter communication, ultimately shaping complex traits. Specifically, we are investigating how the regulatory landscapes of genes mediate the effects of enhancer variation on phenotype. To address these questions, we focus on autoimmunity as a model for complex phenotypes. By integrating comparative and population genomics with advanced computational approaches, we seek to illuminate the mechanisms through which enhancer variation drives phenotypic diversity.